mRNA was considered as a potential anti-tumor vaccination method long before mRNA vaccine against SARS-CoV-2 pushed the technology to center stage, and now, given the overwhelming success of the COVID-19 vaccine, mRNA has been explored for a variety of other applications, including vaccines against other infectious diseases, anti-tumor vaccines, and protein supplementation/replacement therapy, among others. Although mRNA is sensitive to ubiquitous ribonucleases and may be rapidly degraded, it also has strong safety advantages, non-replicating meaning it does not interact with the genome, and its relative instability and rapid metabolic clearance in vivo Can be adjusted by using various modification methods. Existing successful experience has confirmed that after mRNA is incorporated into different delivery structures, rapid uptake and high-efficiency protein expression after administration can be achieved.
The complete mRNA production process faces multiple challenges, including those related to the starting materials (plasmid DNA, pDNA), and those related to the specific properties of mRNA itself, such as its high sensitivity to the nearly ubiquitous ribonuclease and shear stress during processing, and challenges of separation and purification due to the similar size of mRNA to certain impurities such as double-stranded RNA. After obtaining the pDNA template of the desired sequence of interest, the steps for cell-free downstream production of mRNA can be summarized as:
mRNA is synthesized in an enzymatic reaction called in vitro transcription (IVT), in which a template for the target DNA sequence – linearized pDNA, nucleotides (NTPs), and enzymes are mixed together.
The transcribed mRNA is then purified from the reaction mixture/contaminants using steps such as chromatography, tangential flow filtration, and sterile filtration.
The mRNA is finally encapsulated in lipid nanoparticles (LNPs) typically composed of 4 lipid components, and the final product is purified and concentrated using tangential flow filtration (TFF) and/or chromatography before aseptic filling.

Challenges of mRNA manufacturing
mRNA synthesis
Cell-free synthesis of mRNA requires the pDNA template to be linearized with a restriction enzyme that cuts at a specific site of the supercoiled plasmid. Impurities associated with this step need to be removed, such as using tangential flow filtration and/or chromatography. Then, the linearized DNA template is in vitro transcribed (IVT) into mRNA. Add RNA polymerase (such as T7 RNA polymerase) and nucleotide triphosphates to the linearized and purified pDNA. To increase mRNA stability and reduce immunogenicity, some modified nucleotide triphosphates can be used. To stabilize and allow efficient mRNA transduction in cells, a 3' poly-(A) tail and a 5' cap are required. The Poly-(A) tail is critical for terminating protein transcription and translation, preventing 3' exonucleases from digesting mRNA. The 5' cap stabilizes mRNA by preventing degradation by 5' exonucleases. Two methods can be used to cap mRNA. Capping can occur during the transcription step, or, after IVT, and can be achieved enzymatically using vaccinia virus capping enzyme, which generally caps more efficiently but requires purification of the mRNA first.
mRNA purification
After IVT, the transcribed 3'poly-(A)-mRNA-5' cap product needs to be purified from the mixture of endotoxin, immunogenic dsRNA, residual DNA template, RNA polymerase, truncated RNA fragments, unused nucleotide triphosphates and other impurities produced during the capping reaction. Various options are available for the purification of mRNA, typically including one or two chromatography steps, and ultrafiltration/diafiltration steps based on tangential flow filtration.
Affinity chromatography is a commonly used high-efficiency purification strategy, such as Poly (dT) chromatography, which effectively removes DNA, nucleotides, enzymes, buffer components, etc. without poly-(A) by capturing mRNA with poly-A tails, but it cannot distinguish dsRNA from single-stranded RNA (ssRNA). After the initial affinity chromatography, there is usually a second chromatography step, called polishing, to further purify the ssRNA, for example, using anion exchange chromatography. Sometimes a combination of ion exchange and hydrophobic interaction chromatography is used.
An alternative way to separate mRNA from smaller impurities is to use tangential flow filtration with a molecular weight cut-off ranging from 30 to 300 kDa, which also allows concentration and diafiltration in the same unit operation.
In the standard batch-mode IVT process, the desired RNA product is purified from the reaction mixture after the reaction is complete, while the polymerase, pDNA template, and unreacted nucleotide triphosphates are discarded. However, due to the high cost of these IVT reaction components, some developers have attempted to use a fed-batch-like transcription strategy for large-scale mRNA production. During the process, the progress of the IVT reaction was monitored to maintain optimal reaction conditions and components are added to the reactor as needed. The efficiency of obtaining intact full-length RNA products, or the probability of truncated transcripts, depends on the precise control of the RNA sequence and reaction conditions and needs to be optimized to increase process yield.
mRNA encapsulation
RNA is highly sensitive to rapid degradation by ubiquitous ribonucleases, requiring an efficient system that not only stabilizes mRNA but also introduces it into cells. The incorporation of mRNA into the delivery system, along with selected combinations of various lipid components, has been shown to protect mRNA from ribonuclease degradation, enhance cellular uptake, and improve mRNA translation in target cells.
Lipid nanoparticles (LNP) are the most commonly used mRNA delivery system; they are typically composed of 4 different lipid components, including ionizable cationic lipids, helper phospholipids, cholesterol, and polyethylene glycol (PEG)-lipid . Encapsulation of mRNA in lipid complex protects it from ribonuclease degradation while efficiently delivering it into the cytoplasm of cells.
The first step in LNP preparation is to dissolve the lipids in a solvent (e.g. ethanol), then rapidly mix the dissolved lipids with a low pH aqueous buffer containing mRNA using a cross-flow or microfluidic mixer to obtain LNP that encapsulates mRNA, the optimal control of the mixing channel and speed is the key to obtaining the required particle size, particle size distribution, and encapsulation efficiency. The resulting mRNA-LNP complexes were diafiltered to replace the low pH buffer with a neutral buffer. This step should be performed quickly to avoid lipid degradation observed at low pH. Concentration is then performed by ultrafiltration.
Disadvantages of LNPs include that they may require cold chain logistics. Furthermore, due to the particle size of LNP, it is not always possible to perform sterile filtration, in which case alternatives must be considered, such as a fully closed single-use process.
None
 Solutions for mRNA in vitro Synthesis
Solutions for mRNA in vitro Synthesis  Hollow Fiber Filter Module
Hollow Fiber Filter Module 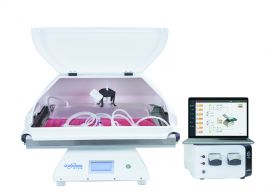 DuoWave® Rocking Single-use Bioreactor
DuoWave® Rocking Single-use Bioreactor  DuoBioX Pro Single-use Bioreactor
DuoBioX Pro Single-use Bioreactor  2D Liquid Storage Bag
2D Liquid Storage Bag  3D Liquid Storage Bag
3D Liquid Storage Bag  Hydrophobic Polytetrafluoroethylene (PTFE) Filter
Hydrophobic Polytetrafluoroethylene (PTFE) Filter  High-Precision Polypropylene Filter
High-Precision Polypropylene Filter  Low-Diffusion Polyethersulfone Filter
Low-Diffusion Polyethersulfone Filter Copyright © Shanghai Duoning Biotechnology Co., Ltd. All Rights Reserved Sitemap | Technical Support: 
Message-